DEEC TALK: Systems Modeling in Single Cells — Consequences of Stochastic Gene Expression
On June 18, the 4th DEEC TALK took place, where Abhyudai Singh Singh, Full Professor at the University of Delaware and PhD in Electrical and Computer Engineering, shared his research, which focuses on the use of mathematical tools applied to biology — particularly the study of cellular behavior. This field of research is currently expanding and benefits from initiatives such as the European Innovation Council (EIC), promoted by the European Commission.
The event began with an introduction by Sérgio Pequito, Vice President for Research, Development, and External Relations.

Throughout the session, Abhyudai Singh Singh discussed the intrinsic variability of gene expression at the single-cell level. He began by addressing cellular decision-making processes: during gene expression, the information encoded in DNA is used to produce proteins through transcription (where the cell creates a complementary copy of DNA — messenger RNA) and translation (where mRNA is “read” to determine the sequence of amino acids needed to form a specific protein).
Based on this gene expression, cells may exhibit higher or lower resistance to drugs, depending on the number and type of proteins produced — a trait that can also be passed on to subsequent generations of cells through division.
However, increases and decreases in protein levels can create transient phases — fluctuations — within the same cell, which may alternate between two distinct states: one of higher resistance and another of lower resistance, due to differences in protein production activity. Singh correlates the time a cell spends in each state with the proportion of cells in that state across the population and with the dynamics of lineage expansion — that is, how quickly the cell gives rise to new generations — while also accounting for stochastic fluctuations.
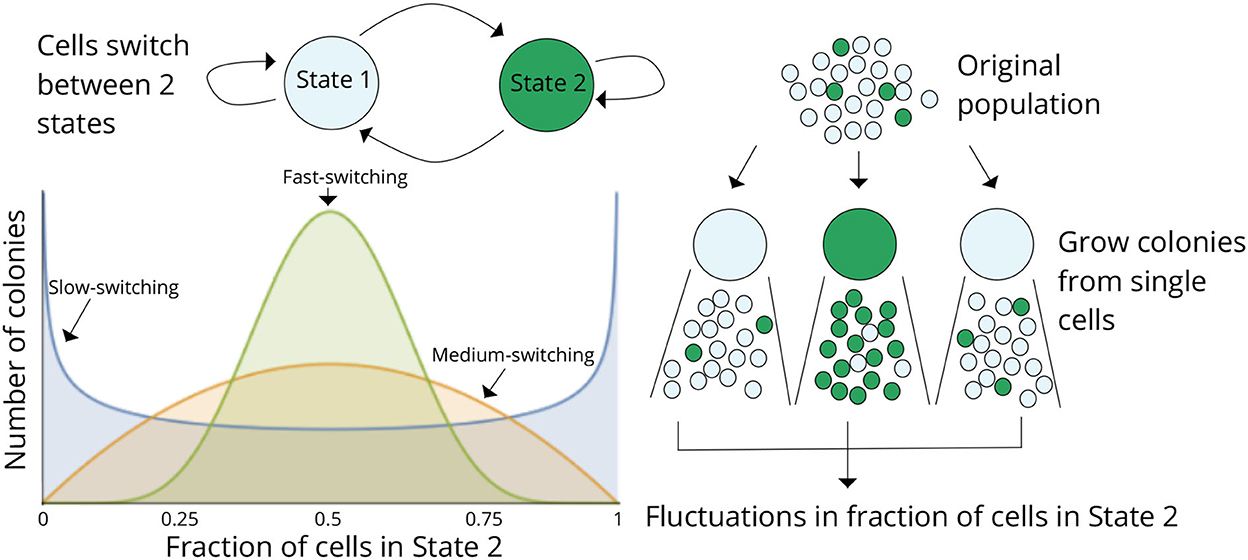
No entanto, ao longo da sua investigação, Abhyudai Singh Singh observou ainda que, ao colocar um vSingh’s research further showed that when a virus is introduced into cells to manipulate gene expression, the lysis period — the time from viral activation to membrane rupture (marking cell death) — remains relatively constant: cells lyse approximately 65 minutes after virus activation, with a deviation of only 3.5 minutes. However, this lysis process introduces additional noise, i.e., variability that complicates the analysis of internal fluctuations.
Em conclusão, o investigador destacou que as flutuações a nível celular são inevitáveis e que podem ser prejuIn conclusion, the researcher emphasized that cellular fluctuations are inevitable and can pose challenges to biological studies. However, mathematical modeling can be used to minimize the noise they introduce. Studying gene expression in this way contributes to a better understanding of biological mechanisms and their associated functions.

The session concluded with a Q&A, where participants had the opportunity to engage directly with the speaker